Reducing Sequence Bias in small-RNA Sequencing
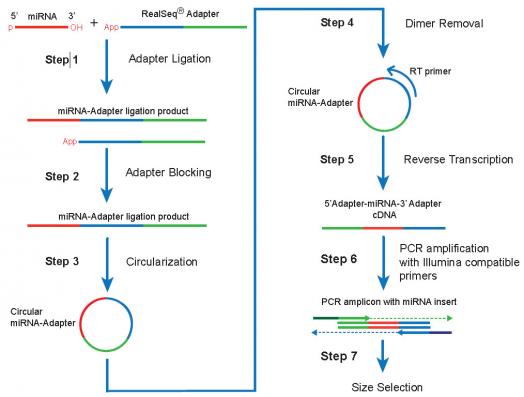
RealSeq®-AC is a novel, proprietary method for preparing small-RNA sequencing libraries that nearly eliminates incorporation bias in Next Generation Sequencing (NSG).
Problem: Commonly used sequencing library preparations lead to underestimation of the abundance of most miRNAs, some by as much as 10,000-fold. This under representation can obscure the presence of some RNAs, including potentially useful biomarkers if they were detected accurately. Most bias originates from sequence-dependent variability in the enzymatic ligation reactions that attach the two adapters to the 3’ and 5’ ends of the miRNAs/small RNAs during preparation of sequencing libraries.
Solution: RealSeq-AC uses a novel single adapter and circularization, greatly reducing library preparation bias.
- Accurately quantifies biologically relevant small RNAs
- Eliminates bias-induced over/under represented small RNAs
- Allows discovery of novel small RNAs
- Efficient and gel-free workflow
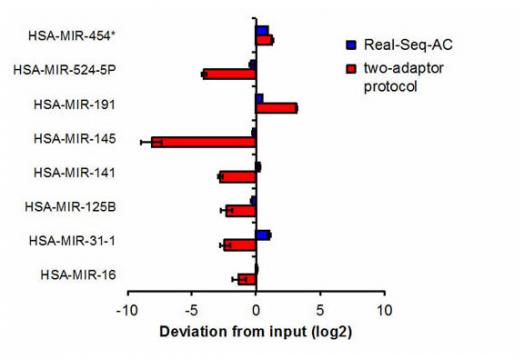
RealSeq-AC reduces sequencing bias in known problematic miRNAs. A commonly used 2-adapter protocol is compared with the RealSeq-AC one-adapter protocol. Deviation from the optimal "0" value are shown for typical under- or over-represented miRNAs. The RealSeq-AC method significantly reduces the detection bias.
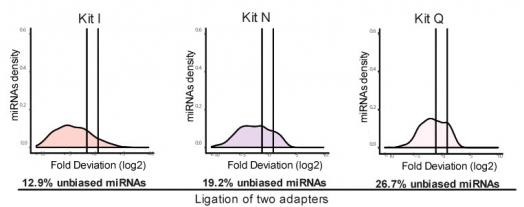
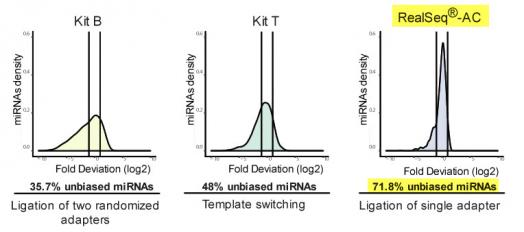
RealSeq-AC reduces incorporation bias compared to other small RNA library preparation kits. All sequencing libraries were prepared in triplicate using 1 pmole of miRXPlore Universal Reference pool (Miltenyi Biotec) which consists of equimolar concentrations of 963 synthetic mature miRNAs. Purified libraries were sequenced on the Illumina MiSeq platform. Trimmed sequencing reads were aligned to a custom miRNA reference (Bowtie2, Langmead et al. 2012). Reads mapping to miRNAs were counted and fold-deviations from the equimolar input were calculated and plotted as log2 values. Measurements of miRNA levels falling within a factor of two of the expected values (between vertical lines) are considered unbiased (Fuchs et al, 2015). The method of adapter attachment to the miRNAs is noted under the graphs.
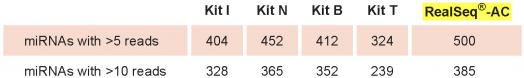
RealSeq-AC detects more miRNAs in total RNA than other miRNA library preparation kits. RealSeq-AC detects more miRNAs with over 5 or 10 reads per million, respectively, from total RNA samples compared to other kits for miRNA sequencing library preparation. RealSeq-AC is optimized for inputs between 1 μg to 100 ng of total RNA.
RealSeq-AC Schematic